Valentines Gummies,Valentine Gummy Candy,Valentine'S Day Gummies,Valentine'S Day Gummy Candy Montreal Shantou Food Co., Ltd , https://www.montrealsnack.com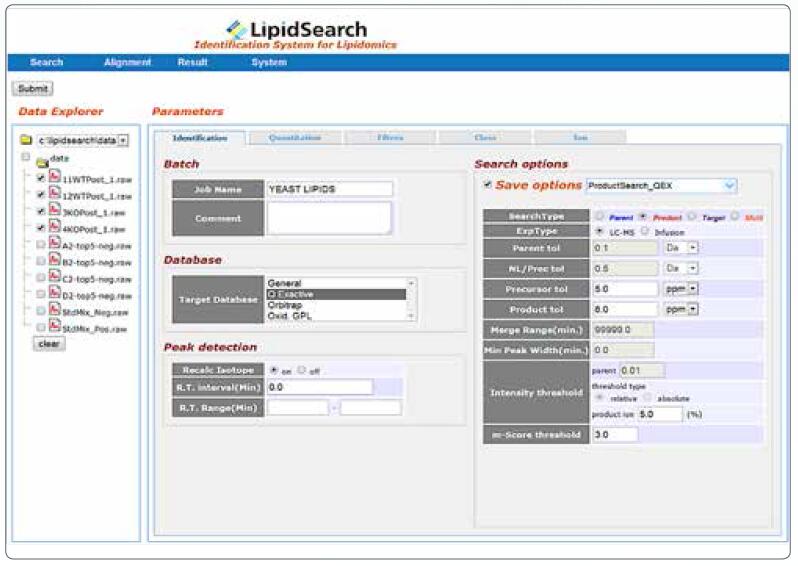



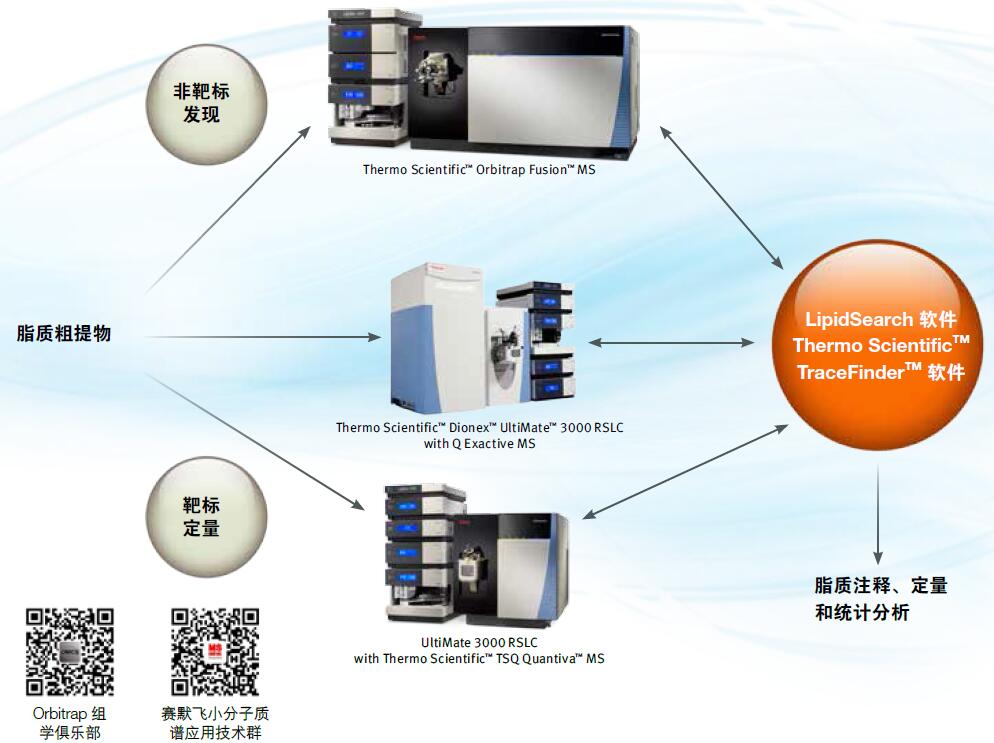
Thermo Scientific LipidSearch software tailored workflow for lipidomics
Automated identification and relative quantification of lipid group LC/MS
Prospects for lipidomics
As an emerging research discipline, lipidomics is important for scientists to understand cell physiology and pathology. The application of lipid profiling in the study of disease phenotype is becoming a direction of vigorous development in translational medicine. Through the identification of specific lipid biomarkers, we hope to distinguish between healthy people and people at risk, to diagnose early diseases, and to promote the establishment of personalized medicine.
LC/MS is an analytical technique widely used in lipidomics analysis. Identification, relative and absolute quantification of lipids in biological samples requires advanced software and a comprehensive database. Thermo ScientificTM LipidSearchTM software provides accurate lipid identification of LC/MS data and automatically integrates a streamlined summary report of complex data output. The software operation interface is web-based and easy to use, which greatly reduces the time required for data analysis.
Easy-to-use web browser interface for reliable, automated authentication
LipidSearch software for automatic lipid identification and relative quantitative analysis
LipidSearch software was jointly developed by Professor Ryo Taguchi and MKI (Tokyo, Japan). The software is powerful enough to analyze large amounts of data collected in nano-infusion or LC/MS experiments for automated identification and relative quantification of lipid molecules. The industry-leading high-resolution, precision mass spectrometer, Thermo Scientific Orbitrap, and the exclusive LipidSearch software combine to deliver the most accurate and reliable lipid profiling and identification results at your fingertips.
• Compatible with experimental data collected by Thermo ScientificTM triple quadrupole, ion trap and Orbitrap mass spectrometer
• Equipped with the largest lipid group database containing over 1.7 million lipid ions and their predicted fragment ions
• Provides different lipid identification algorithms for daughter ions, precursor ions, and neutral loss scans
• Ability to correlate lipid data obtained from multiple LC/MS and MSn experiments
• Ability to quantify precursor ions for lipid identification in LC/MS or direct injection experiments
The database contains defined molecular structures and covers more than 1.7 million lipid ions and their predicted fragment ion information . The calculation and optimization of the ion fragmentation mode is based on experimental results and professional experience. Fingerprint information for lipid adduct ions and MSn is included in the database. All data is stored in an XML file format and is customizable.
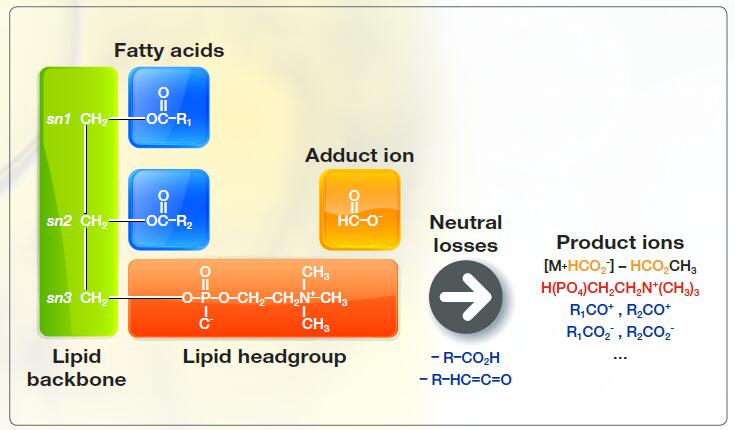
Fragment ions corresponding to lipid heads, fatty acids, and backbone
LipidSearch software gives you an easy-to-use automated workflow
From peak extraction, relative quantification to identification, LipidSearch software gives you an easy-to-use, automated workflow.
Step 1
Data Analysis Module - Peak Extraction
• Raw data file read
• Intelligent peak extraction
The peak extraction engine integrated into LipidSearch software handles data collected by different MS experiments and instrumentation platforms. Combine unique peak extraction algorithms, distinguish between different experiments and instrument types, and integrate advanced spectral processing functions to ensure accurate peak extraction.
Step 2
Identification Module - Lipid Identification
• Specific lipid class identification (targeting)
• Comprehensive comprehensive identification (non-targeting)
• Score algorithm
LipidSearch software provides two different authentication and corresponding scoring algorithms:
• Specific lipid class algorithms are used to analyze data from precursor ion scans and neutral loss scans, and to identify lipids by polar head class or fatty acid information.
• A comprehensive comprehensive identification algorithm for analyzing daughter ion scan data to identify lipids by matching against predicted lipid fragmentation patterns in the database.
• LipidSearch software provides a range of scoring algorithms to achieve reliable results.
Step 3
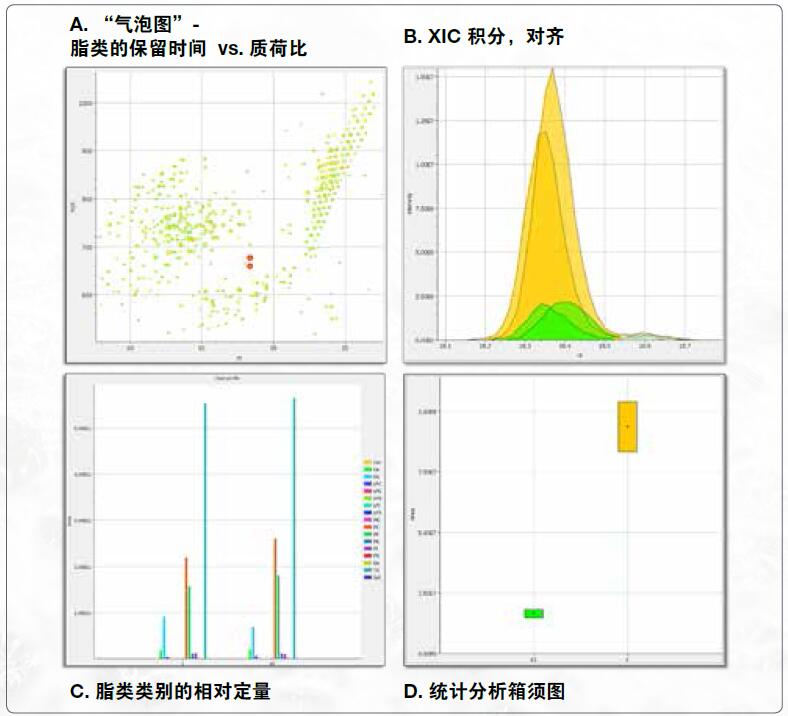
Quantitative module - alignment and quantification
• XIC peak area integral
• Retention time alignment
• Relative quantification
• Statistical analysis
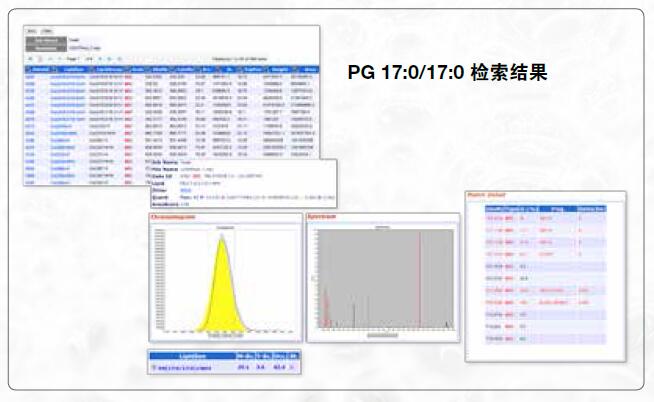
Prior to the quantitative analysis, the lipid identification results for each sample were aligned within a retention time deviation window. The quantification of lipids is achieved by integrating the extracted ion chromatogram of the precursor ions in the full-sweep spectrum. To accurately calculate the peak area, the chromatographic peaks are first denoised and smoothed, then the partially overlapping peaks are separated and finally calculated. The statistical difference between the control group and the control group was compared by t test, and the average result of each group of peak areas was shown by the "box whisker".
CoQ 7 , [M+NH4] + alignment result
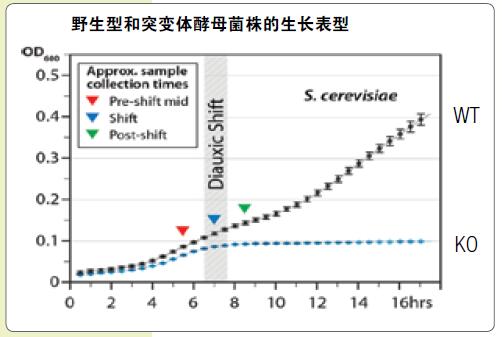
Lipid profile analysis of wild-type and mutant yeast strains reveals subtle changes in lipids
The wild type (WT) yeast (S. cerevisiae) can continue to grow after the consumption of glucose in the medium (diuxic shift point), while the mutant (KO) strain cannot be defective due to CoQ synthesis. Continue to grow. Samples of wild-type and mutant strains after the two-phase growth point were taken and subjected to lipid group analysis in MS and data-dependent MS2 mode using a Thermo ScientificTM Q ExactiveTM quadrupole-orbit trap mass spectrometer.
step 1
data processing
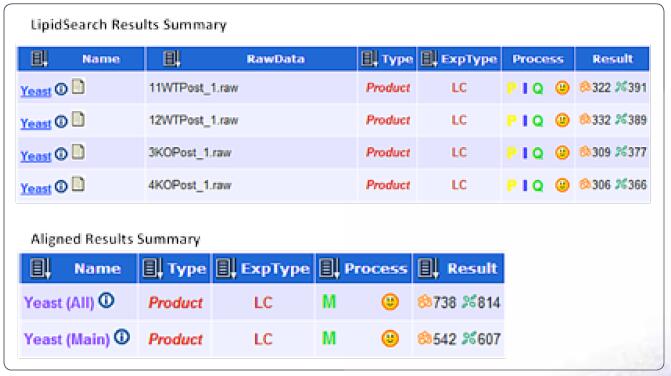
Search for possible lipid compounds in LC/MS raw files containing MS and data-dependent MS2 spectral data, including FA (fatty acid), sphingosine, Lyso-GP (lysophospholipid), MG (monoalkylglycerol) , GP (PA, PC, PE, PG, PI, PS), Cer (ceramide) and CoQ (coenzyme). Retrieve the quality deviation setting: the precursor ion mass deviation window is set to 5 ppm and the daughter ion is set to 8 ppm. All sample identification results were aligned within a 0.25 min retention time window and a comprehensive report was generated. In the data file of this experiment, a total of 738 lipids and isomers were identified, corresponding to 542 different molecular formulas.
LC/MS chromatogram of yeast lipids
LipidSearch status window view for search and alignment results
Step 2
Identification
Summarize the results of each MS2 spectrum and give a score that matches the predicted fragmentation pattern in the database. If a mixture of lipids is found, the most abundant lipid results are shown. When the results are selected, the fragment ion peaks used to participate in the identification are highlighted in red.
ID of PE 16:0-18:1, MS2 of m/z 716.5230 [C39H75NO8P]-
Step 3
Alignment and quantification
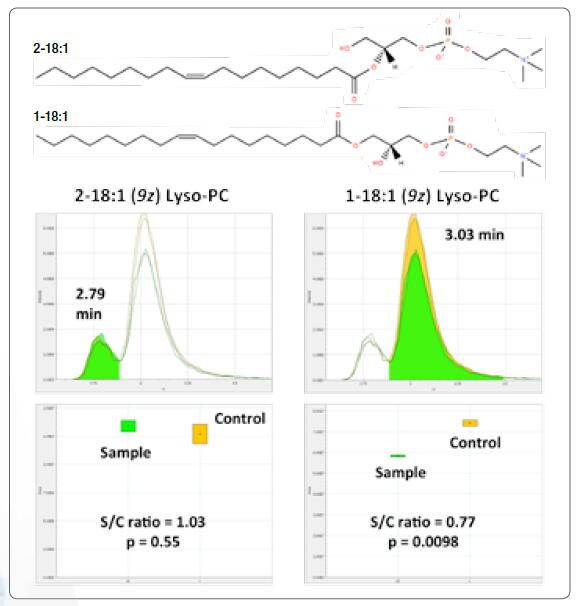
Summarize the results of each MS2 spectrum and give a score that matches the predicted fragmentation pattern in the database. If a mixture of lipids is found, the most abundant lipid results are shown. When the results are selected, the fragment ion peaks used to participate in the identification are highlighted in red. Align and summarize the same lipids eluted within a specific retention time deviation window. Report relative peak area and t-test statistical analysis results. The alignment chromatograms of the two Lyso-PC 18:1 isomers are shown on the right, and the corresponding structure can be determined based on the elution order of the lysophospholipids on a reverse phase HPLC column.
By using the industry-leading high-resolution, precision-quality Thermo ScientificTM OrbitrapTM technology combined with the exclusive LipidSearch professional software, you can easily achieve the most accurate and reliable lipid profile and lipid identification analysis at an unprecedented rate. LipidSearch software is also available for the ThermoScientific TSQ Triple Quadrupole Mass Spectrometry System, which supports targeted lipid quantification. For both lipid group analysis workflows, LipidSearch software automates the analysis of complex data and generates comprehensive reports to help you greatly improve the efficiency of your data analysis.
Minimum/Recommended PC Requirements
OS: Microsoft® Windows® 7 Professional (x64) or Windows 8 (x64); English language
CPU: Quad - or multi-core CPU, 3 GHz or higher
Memory: 16 GB RAM or higher
HDD: At least 500 GB free space for LipidSearch software and data
At least 2 TB HDD for OS/software or larger for storage
SSD (optional): At least 50 GB free space for LipidSearch software and data
At least 500 GB solid-state drive for OS/software or larger